Biological structures
Posted on 09/01/2014

Brett Collins
Biology on the edge – Adaptor Protein complex 2
What does it look like?
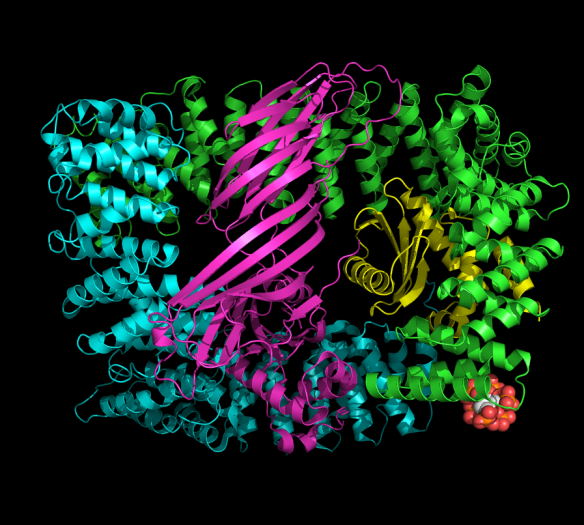
The image was generated using the molecular graphics software PyMOL. http://www.pymol.org/
Cartoon representation of the core subunits of the AP2 adaptor complex (PDB ID 2VGL, Collins et al., Cell, 2002). The four protein subunits of the complex are shown in ribbon representation and the bound inositolhexakisphosphate (IP6) molecule is shown in spheres. IP6 mimics the lipid head group required for attaching the protein to the cell membrane.
What is it?
Every minute, almost every cell in our body forms thousands of small membrane-bound vesicles at their surface. These tiny structures are about 100 nm (thats 1 tenth of 1 millionth or 0.0000001 metres) in diameter and are essential for cells to ingest nutrients, hormones and other molecules through a process called endocytosis. These vesicles are formed via a complex ballet of molecular interactions between membrane lipids and protein molecules. Two protein complexes are absolutely essential – clathrin and Adaptor Protein complex 2, or AP2 for short. AP2 is a very large assembly and contains four different proteins, or subunits, that are tightly held together by hydrophobic interactions, hydrogen bonds and electrostatic interactions. AP2 binds membrane lipids called phosphoinositides which pull it to the cell surface, and it is then able to undergo a dramatic structural change to bind transmembrane receptors and recruit them into vesicles for cell uptake. This process of membrane trafficking is one of the fundamental differences between more complex eukaryotic cells and simpler bacterial species.
Where did the structure come from?
This was the first structure Brett Collins solved when he joined the lab of David Owen in Cambridge fresh out of his PhD. The structure was solved using X-ray diffraction with data collected at the European Synchrotron Radiation Facility (ESRF) in Grenoble, France. You can view it at http://www.rcsb.org/pdb/explore.do?structureId=2VGL and read more about it in http://www.ncbi.nlm.nih.gov/pubmed/12086608.
Related articles
|
Munc18: No Munc-eying around with our molecular transport machinery! |
The structure of empty space – HKUST 1 |
Magnetic monopoles in the pyrochlore structure |