DNA Polymerase in Celebration of Evolution Day
Today Anton tells us about a structure behind one aspect of evolution.
What does it look like?
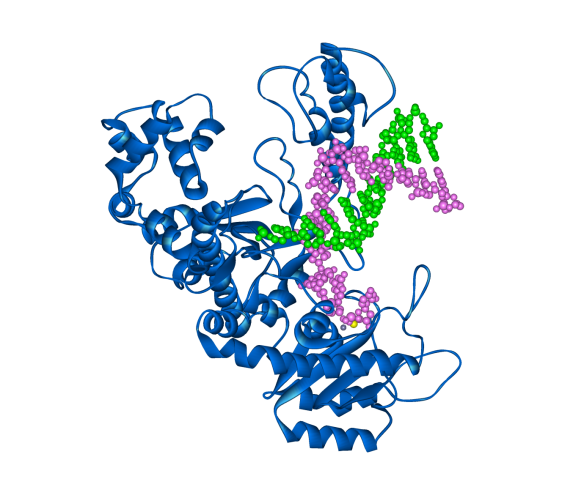
The Klenow fragment of DNA polymerase I from E. coli (in blue) with template and new DNA strands (green and magenta respectively).
What is it?
Today is Evolution Day, the day when the publication of Charles Darwin's seminal work On the Origin of Species in 1859 is celebrated. DNA and its specific sequence of bases: A, T, C, and G, have a lot to do with evolution, but since we've already blogged about the structure of DNA for Rosalind Franklin's birthday, I thought I'd write about the protein that replicates the DNA when a cell divides: DNA polymerase.
DNA replication is semi-conservative: that is, each strand of DNA acts as the template for the new copy and one of each copy goes to the two new daughter cells. There are billions of A, T, C, and G's that make up a genome; thus it is of the utmost importance that DNA polymerase is extremely accurate in its job. Typically DNA polymerase will make one mistake for every billon bases copied. There are a number of reasons why DNA polymerase is so accurate.
- DNA polymerase can select the correct bases for the correct pairing (i.e. A pairs with T and G pairs with C).
- There is a proof reading mechanism.
- Repair systems if a mistake is spotted.
Many of those one in a billion mistakes will never show as a unique characteristic, but some could result in a random mutation that is seen as observable characteristics (the phenotype) in an organism. If that mutation is in the reproductive cells then the mutation could be passed onto the next generation and, with the favour of natural selection, propagate for many generations.
As a mistake in copying DNA can prove to be disastrous as well as beneficial there is a lot of redundancy in DNA replication, with many organisms having more than one DNA polymerase. E. coli for instance has five DNA polymerases. The structure shown today is the Klenow fragment of E. coli DNA polymerase I. The Klenow fragment is the part of the polymerase where replication and proof reading occur but other domains have been removed. The structure resembles the shape of a hand with the DNA being held between a finger and thumb.
Where did it come from?
DNA polymerase I from E. coli was discovered by Arthur Kornberg in 1956 (and later won the Nobel Prize for Physiology and Medicine in 1959 for the discovery). The structure shown here was solved in 1993 [1] by Lorena Beese, Victoria Derbyshire, and Thomas Steitz (who won the Nobel Prize for Chemistry in 2009 with Venkatraman Ramakrishnan and Ada Yonath for their work on the ribosome). The structure was taken from the Protein Data Bank (PDB number: 1KLN).
[1] L. S. Beese, V. Derbyshire, T. A. Steitz (1993). Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science 260: 352-355.