Chymotrypsin — the first protease
Today we have a guest post from Professor Stephen Curry, Professor of Structural Biology at Imperial College London. Stephen blogs at Occam's Typewriter and the Guardian and was one of the founding members of the Science is Vital UK campaign group. He is currently visiting Australia and is giving a series of public lectures, the next in Sydney on 18th August (2014).

Without further ado – here is Stephen's post.
What does it look like?
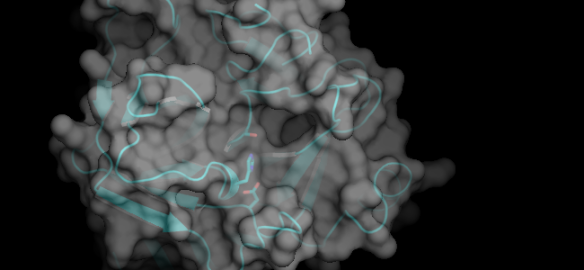
This is a schematic representation of the protein chymotrypsin.
This image, made with PyMOL (using the coordinates from the Protein Data Bank entry 4CHA), shows the overall fold of the polypeptide chain of the molecule, along with a semi-transparent rendering of its surface. The lumpiness of the surface is due to the individual atoms of the protein because the structure has been determined at very high resolution.
What is it?
Chymotrypsin a protease molecule that participates in the digestion in the small intestine of proteins consumed as food. It acts like a molecular pair of scissors but instead of having sharp blades for cutting chymotrypsin uses chemistry. What this means is that it catalyses the cleavage of the peptide bonds between amino acids in selected targets. The three amino acid side-chains of the catalytic centre are shown as sticks in the picture above — from the top these are serine, histidine and aspartic acid. To the right of them you can see a deep pocket that chymotrypsin uses to grab hold of a large, aromatic side-chain in a target protein. During this transient embrace, the Ser-His-Asp 'catalytic triad' works, along with a water molecule, to destabilise and then break the adjacent peptide bond in the target. Repeated action of chymotrypsin and other proteases in the digestive tract breaks proteins down to their component amino acids so that ultimately they can be used by the body to build up new proteins, or as the starting material for synthesizing other essential molecules.
Where did the structure come from?
The structure of chymotrypsin — published initially by a team led by David Blow — was only the fourth protein structure to be solved by X-ray crystallography. It is a landmark because it was the first protease structure to be worked out by the technique and almost immediately revealed not just the architecture of the polypeptide chain but the detailed chemical mechanism that it deployed to break peptide bonds. The Ser-His-Asp catalytic triad has since been found in many other proteases in related and unrelated structures, providing intriguing molecular examples of convergent and divergent evolution.
Personally, the structure has a special meaning since David Blow was an inspirational figure who helped me to get into protein crystallography at the outset of my career in 1989. I was fortunate to be able to return the favour many years later when my group solved the structure of the 3C protease from foot-and-mouth disease virus which turns out to be a distant relative of chymotrypsin.