A different type of donut – beta-sliding clamp
What does it look like?
Beta has a striking resemblance to a donut, but isn't as tasty. The protein is actually a dimer, with the arrows showing the dimeric interface. Each dimer has three domains comprised of two beta-sheets and two alpha-helices.
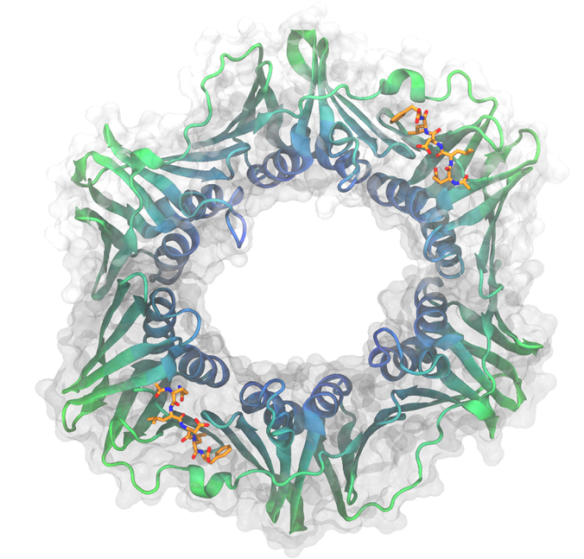
Image generated by VMD (Visual Molecular Dynamics) using the coordinates from the Protein Data Bank (PDB code: 3Q4J)
What is it?
The Escherichia coli beta-sliding clamp (beta) is an aptly named DNA replication protein. It is clamped onto and slides along DNA, recruiting other replication proteins to the DNA during the different stages of replication. What makes this an interesting protein is the nature of these interactions, as every protein that interacts with beta binds in the same pocket by very similar amino acid motifs. On the structure, the consensus amino acid motif (QLDLF) is bound into the pocket. This conserved binding has made the E. coli beta protein a target for novel antibiotics, and many potential antibacterial small molecules have been published.
Where does it come from?
The first crystal structure was solved by X.P. Kong and colleagues in 1992 [1] and since then, many structures of E. coli beta in complex with protein binding partner consensus motifs [2] and small molecule inhibitors [3] have been solved.
[1] Kong, X.P., Onrust, R., O’Donnell, M., and Kuriyan, J. (1992). Three-dimensional structure of the b subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell 69(3): 425-437
[2] Wolff, P., Olieric, V., Briand, J.P., Chaloin, O., Dejeagere, A., Dumas, P., Ennifar, E., Guichard, G., Wagner, J., and Burnouf, D.Y. (2011). Structure-based design of short peptide ligands binding onto the E. coli processivity ring. Journal of Medicinal Chemistry 54(13): 4627-4637
[3] Yin, Z., Whittell, L.R., Wang, Y., Jergic, S., Lui, M., Harry, E.J., Dixon, N.E., Beck, J.L., Kelso, M.J., and Oakley, A.J. (2014). Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach. Journal of Medicinal Chemistry 57: 2799-2806