Brace yourself! The structure of DiSulfide Bond proteins (DSBs)
What does it look like?
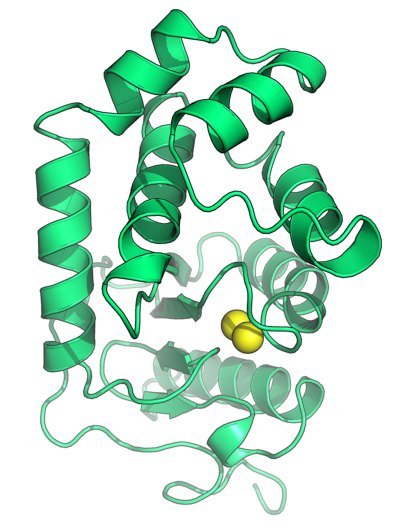
The image was generated using the molecular graphics software PyMOL. http://www.pymol.org/
Cartoon representation of the disulfide oxidoreductase DsbA from Burkholderia pseudomallei (PDB ID 4K2D, Ireland et al., Antioxid Redox Signal, 2013, epub doi:10.1089/ars.2013.5375). The sulfur atoms of the catalytic cysteines are shown as yellow spheres.
What is it?
The world outside can be a hostile place – particularly when you’re just a bug.
Masters of survival, bacteria deploy an arsenal of proteins, integrated in their surface and secreted extracellularly, that allow them to colonize environments peppered with digestive enzymes, unfavourable pH and temperature conditions, and a variety of other environmental insults.
To resist such an onslaught, disulfide bonds – a type of chemical linkage that adds structural bracing – reinforce many of the bacterial proteins that are exposed to the extracellular environment. These disulfide bonds are introduced by the co-operative action of a family of protein folding enzymes called DiSulfide Bond proteins (DSBs). Pictured is a DSB (DsbA) from the pathogen Burkholderia pseudomallei, a soil-dwelling bacterium endemic in SE Asia and Northern Australia that causes the human disease melioidosis. B. pseudomallei lacking DsbA are unable to establish lethal infection in mice, reflecting the pivotal role that this enzyme plays in the folding and functioning of many bacterial proteins needed for infection. For this reason, B. pseudomallei DsbA is a promising target for antibacterial inhibitor development and its structure provides the starting point for future drug design efforts.
Where did the structure come from?
This structure was determined by X-ray diffraction methods at the Australian Synchrotron. You can view it at http://www.rcsb.org/pdb/explore/explore.do?structureId=4K2D and read more about the role of DsbA in B. pseudomallei virulence at http://www.ncbi.nlm.nih.gov/pubmed/23901809.